aplms fits additive partial linear models with autoregressive symmetric errors.
This method is suitable for data sets where the response variable is continuous and symmetric,
with either heavy or light tails, and measured over time.
The model includes a parametric component for a set of covariates, while another set of covariates
can be specified as semi-parametric functions, typically time-related.
In this setup, natural cubic splines or cubic P-splines are used to approximate the nonparametric components.
Arguments
- formula
A symbolic description of the parametric component of the model to be fitted. The details of model specification are given under ‘Details’.
- npc
A vector of names of non parametric component.
- basis
A vector of names of the basis to be used for each non parametric covariate.
- Knot
A vector of the number of knots in each non-linear component of the model.
- data
A data frame containing the variables in the model
- family
Symmetric error distribution. The implemented distribution are:
Normal(),LogisI(),LogisII(),Student(df),Powerexp(k),Gstudent(parm=c(s,r)).- p
autoregressive order of the error
- control
optimization rutine.
- init
A list of initial values for the symmetric error scale, phi, and autoregressive coefficients, rhos.
- lam
smoothing parameter vector.
- verbose
Logical; if TRUE, prints estimation progress messages. Default is FALSE.
- x
APLMS object.
- ...
other arguments
Value
Returns an object of class “aplms”, a list with following components.
- formula
the
formulaobject used.- family
the
familyobject used.- npc
the
npcobject used.- Knot
the
Knotobject used.- lam
the
lamobject used.- rdf
Degrees of freedom:
n - q - p - 1.- VAR_F
Estimate the asymptotic covariance matrix for the gamma parameters.
- basis
The
basisto be used for each non parametric covariate.- WALD_f
The summary table of the Wald statistics.
- summary_table_phirho
The summary table of the rho and phi parameters.
- N_i
Basis functions.
- f
Estimated gamma parameters.
- Dv
Dv values for the symmetric error.
- Dm
Dm values for the symmetric error.
- Dc
Dc values for the symmetric error.
- Dd
Dd values for the symmetric error.
- delta
delta_i for the symmetric error.
- LL_obs
Observed information matrix of the fitted model.
- loglike
The estimated loglikelihood function of the fitted model.
- total_df
The total effective degree of freedom of the model.
- parametric_df
The degree of freedom of the parametric components.
- npc_df
The effective degree of freedom of the non parametric components.
- AIC
Akaike information criterion of the estimated model.
- BIC
Bayesian information criterion of the estimated model.
- AICC
Corrected Akaike information criterion of the estimated model.
- GCV
The generalized cross-validation (GCV).
- yhat
The fitted response values of the model.
- muhat
The fitted mean values of the model.
- residuals_y
The response residuals
- residuals_mu
Raw (Ordinary) residuals: \(y_t - (\textbf{x}_i^\top\beta + f_1(t_{i1}) + \ldots + f_k(t_{ik}))\)
- data
the
dataobject used.- this.call
the function call used.
References
Chou-Chen, S.W., Oliveira, R.A., Raicher, I., Gilberto A. Paula (2024) Additive partial linear models with autoregressive symmetric errors and its application to the hospitalizations for respiratory diseases Stat Papers 65, 5145–5166. doi:10.1007/s00362-024-01590-w
Oliveira, R.A., Paula, G.A. (2021) Additive partial linear models with autoregressive symmetric errors and its application to the hospitalizations for respiratory diseases Comput Stat 36, 2435–2466. doi:10.1007/s00180-021-01106-2
Examples
data(temperature)
temperature.df = data.frame(temperature,time=1:length(temperature))
model<-aplms::aplms(temperature ~ 1,
npc=c("time"), basis=c("cr"),Knot=c(60),
data=temperature.df,family=Powerexp(k=0.3),p=1,
control = list(tol = 0.001,
algorithm1 = c("P-GAM"),
algorithm2 = c("BFGS"),
Maxiter1 = 20,
Maxiter2 = 25),
lam=c(10))
plot(model)
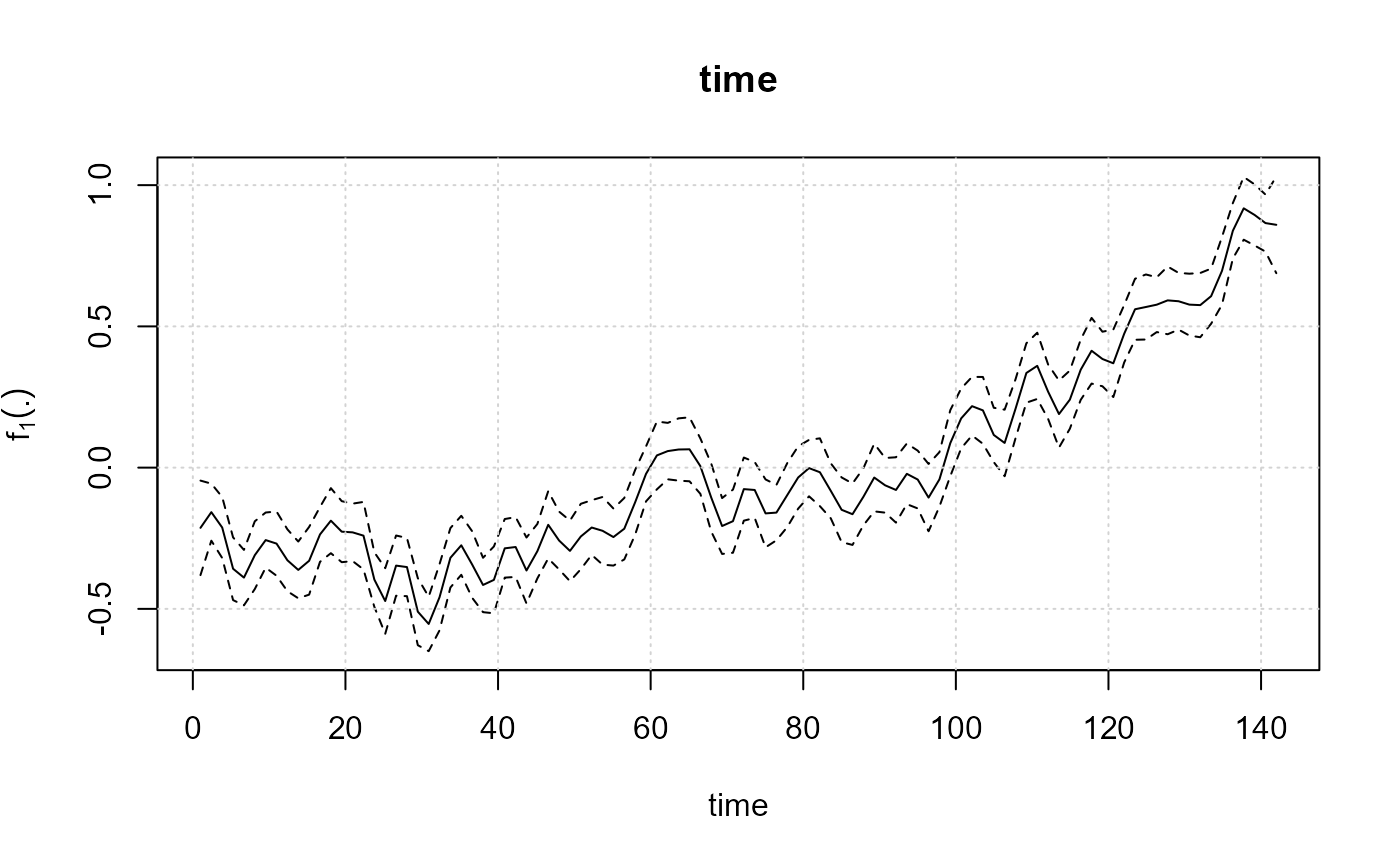 #> [[1]]
#> cov fmean fmean_ls fmean_li
#> 1 1.000000 -0.213215062 -0.38051702 -0.045913101
#> 2 2.424242 -0.157657300 -0.25860288 -0.056711725
#> 3 3.848485 -0.212090507 -0.32001771 -0.104163305
#> 4 5.272727 -0.357959506 -0.46898184 -0.246937174
#> 5 6.696970 -0.389381087 -0.48749987 -0.291262301
#> 6 8.121212 -0.309883662 -0.42979400 -0.189973326
#> 7 9.545455 -0.256507180 -0.35408994 -0.158924423
#> 8 10.969697 -0.268972025 -0.38269634 -0.155247707
#> 9 12.393939 -0.328028685 -0.43751976 -0.218537614
#> 10 13.818182 -0.362119686 -0.46252234 -0.261717030
#> 11 15.242424 -0.329739301 -0.44963766 -0.209840943
#> 12 16.666667 -0.236456854 -0.33345593 -0.139457776
#> 13 18.090909 -0.187783201 -0.30292439 -0.072642015
#> 14 19.515152 -0.226748006 -0.33476704 -0.118728976
#> 15 20.939394 -0.229175092 -0.33080619 -0.127543993
#> 16 22.363636 -0.240824195 -0.36025925 -0.121389135
#> 17 23.787879 -0.396329451 -0.49288877 -0.299770132
#> 18 25.212121 -0.472299770 -0.58853860 -0.356060937
#> 19 26.636364 -0.346859356 -0.45344989 -0.240268819
#> 20 28.060606 -0.352051750 -0.45504127 -0.249062233
#> 21 29.484848 -0.509917727 -0.62863393 -0.391201528
#> 22 30.909091 -0.553447782 -0.64976105 -0.457134516
#> 23 32.333333 -0.458669461 -0.57595336 -0.341385561
#> 24 33.757576 -0.319163400 -0.42430868 -0.214018119
#> 25 35.181818 -0.274962044 -0.37937956 -0.170544531
#> 26 36.606061 -0.343014587 -0.46081001 -0.225219168
#> 27 38.030303 -0.415401411 -0.51166773 -0.319135092
#> 28 39.454545 -0.397422379 -0.51569997 -0.279144793
#> 29 40.878788 -0.285716394 -0.38941287 -0.182019919
#> 30 42.303030 -0.281039407 -0.38691070 -0.175168109
#> 31 43.727273 -0.364210514 -0.48097224 -0.247448788
#> 32 45.151515 -0.295850803 -0.39226072 -0.199440886
#> 33 46.575758 -0.202393200 -0.32149651 -0.083289890
#> 34 48.000000 -0.258064413 -0.36036400 -0.155764823
#> 35 49.424242 -0.294762948 -0.40206745 -0.187458442
#> 36 50.848485 -0.243743271 -0.35944671 -0.128039829
#> 37 52.272727 -0.212056003 -0.30881412 -0.115297886
#> 38 53.696970 -0.223624912 -0.34332922 -0.103920599
#> 39 55.121212 -0.245821845 -0.34682625 -0.144817439
#> 40 56.545455 -0.216226869 -0.32497022 -0.107483516
#> 41 57.969697 -0.122563126 -0.23710176 -0.008024492
#> 42 59.393939 -0.022064966 -0.11936732 0.075237389
#> 43 60.818182 0.043246936 -0.07680916 0.163303035
#> 44 62.242424 0.058536017 -0.04133763 0.158409663
#> 45 63.666667 0.064177908 -0.04607577 0.174431583
#> 46 65.090909 0.064875182 -0.04834389 0.178094259
#> 47 66.515152 0.005296521 -0.09272732 0.103320363
#> 48 67.939394 -0.105963819 -0.22614647 0.014218833
#> 49 69.363636 -0.206727445 -0.30561597 -0.107838917
#> 50 70.787879 -0.189575482 -0.30134938 -0.077801580
#> 51 72.212121 -0.076046167 -0.18782007 0.035727734
#> 52 73.636364 -0.079103697 -0.17799222 0.019784831
#> 53 75.060606 -0.162040947 -0.28222360 -0.041858296
#> 54 76.484848 -0.159214768 -0.25723861 -0.061190926
#> 55 77.909091 -0.096978598 -0.21019767 0.016240479
#> 56 79.333333 -0.034892264 -0.14514594 0.075361411
#> 57 80.757576 -0.001654978 -0.10152862 0.098218668
#> 58 82.181818 -0.016382605 -0.13643870 0.103673494
#> 59 83.606061 -0.082110591 -0.17941295 0.015191763
#> 60 85.030303 -0.149307173 -0.26384581 -0.034768540
#> 61 86.454545 -0.164889347 -0.27363270 -0.056145994
#> 62 87.878788 -0.103394785 -0.20439919 -0.002390379
#> 63 89.303030 -0.035126676 -0.15483099 0.084577636
#> 64 90.727273 -0.062581265 -0.15933938 0.034176852
#> 65 92.151515 -0.079086931 -0.19479037 0.036616512
#> 66 93.575758 -0.021958536 -0.12926304 0.085345970
#> 67 95.000000 -0.042156899 -0.14445649 0.060142691
#> 68 96.424242 -0.106105596 -0.22520891 0.012997714
#> 69 97.848485 -0.041483460 -0.13789338 0.054926457
#> 70 99.272727 0.086897127 -0.02986460 0.203658853
#> 71 100.696970 0.174316765 0.06844547 0.280188063
#> 72 102.121212 0.217565731 0.11386926 0.321262206
#> 73 103.545455 0.202671643 0.08439406 0.320949227
#> 74 104.969697 0.115659933 0.01939361 0.211926256
#> 75 106.393939 0.087320078 -0.03047534 0.205115498
#> 76 107.818182 0.208210642 0.10379313 0.312628152
#> 77 109.242424 0.335094675 0.22994939 0.440239956
#> 78 110.666667 0.360344269 0.24306036 0.477628175
#> 79 112.090909 0.268794480 0.17248123 0.365107728
#> 80 113.515152 0.189627094 0.07091091 0.308343273
#> 81 114.939394 0.240639673 0.13765014 0.343629210
#> 82 116.363636 0.346623508 0.24003297 0.453214045
#> 83 117.787879 0.413779514 0.29754069 0.530018341
#> 84 119.212121 0.384577682 0.28801824 0.481137123
#> 85 120.636364 0.369334466 0.24989923 0.488769704
#> 86 122.060606 0.473909701 0.37227863 0.575540771
#> 87 123.484848 0.560667717 0.45264868 0.668686751
#> 88 124.909091 0.568661309 0.45351981 0.683802810
#> 89 126.333333 0.576904017 0.47990550 0.673902532
#> 90 127.757576 0.592069051 0.47217142 0.711966684
#> 91 129.181818 0.589034350 0.48862971 0.689438988
#> 92 130.606061 0.577041729 0.46755078 0.686532679
#> 93 132.030303 0.575296888 0.46156731 0.689026471
#> 94 133.454545 0.606978315 0.50938891 0.704567721
#> 95 134.878788 0.696587825 0.57665900 0.816516648
#> 96 136.303030 0.838493153 0.74033856 0.936647746
#> 97 137.727273 0.917933747 0.80690816 1.028959338
#> 98 139.151515 0.894514412 0.78638554 1.002643279
#> 99 140.575758 0.865726591 0.76474733 0.966705850
#> 100 142.000000 0.859592517 0.68831338 1.030871657
#>
summary(model)
#> ---------------------------------------------------------------
#> Additive partial linear models with symmetric errors
#> ---------------------------------------------------------------
#> Sample size: 142
#> -------------------------- Model ---------------------------
#>
#> aplms::aplms(formula = temperature ~ 1, npc = c("time"), basis = c("cr"),
#> Knot = c(60), data = temperature.df, family = Powerexp(k = 0.3),
#> p = 1, control = list(tol = 0.001, algorithm1 = c("P-GAM"),
#> algorithm2 = c("BFGS"), Maxiter1 = 20, Maxiter2 = 25),
#> lam = c(10))
#>
#> ------------------- Parametric component -------------------
#>
#> Estimate Std. Error t value Pr(>|t|)
#> intercept 0.056619 0.0041 13.8905 < 2.2e-16 ***
#>
#> ----------------- Non-parametric component ------------------
#>
#> Wald df Pr(>.)
#> time 7589.838 58.583 < 2.2e-16 ***
#>
#> --------------- Autoregressive and Scale parameter ----------------
#>
#> Estimate Std. Error Wald Pr(>|t|)
#> phi 0.0022992 0.0003 7.3902 1.242e-11 ***
#> rho1 -0.2571215 0.0662 -3.8866 0.0001568 ***
#>
#>
#> ------ Penalized Log-likelihood and Information criterion------
#>
#> Log-lik: 200.07
#> AIC : -276.97
#> AICc : -274.46
#> BIC : -94.94
#> GCV : 0.01
#>
#> --------------------------------------------------------------------
print(model)
#> Call:
#> aplms::aplms(formula = temperature ~ 1, npc = c("time"), basis = c("cr"),
#> Knot = c(60), data = temperature.df, family = Powerexp(k = 0.3),
#> p = 1, control = list(tol = 0.001, algorithm1 = c("P-GAM"),
#> algorithm2 = c("BFGS"), Maxiter1 = 20, Maxiter2 = 25),
#> lam = c(10))
#> [[1]]
#> cov fmean fmean_ls fmean_li
#> 1 1.000000 -0.213215062 -0.38051702 -0.045913101
#> 2 2.424242 -0.157657300 -0.25860288 -0.056711725
#> 3 3.848485 -0.212090507 -0.32001771 -0.104163305
#> 4 5.272727 -0.357959506 -0.46898184 -0.246937174
#> 5 6.696970 -0.389381087 -0.48749987 -0.291262301
#> 6 8.121212 -0.309883662 -0.42979400 -0.189973326
#> 7 9.545455 -0.256507180 -0.35408994 -0.158924423
#> 8 10.969697 -0.268972025 -0.38269634 -0.155247707
#> 9 12.393939 -0.328028685 -0.43751976 -0.218537614
#> 10 13.818182 -0.362119686 -0.46252234 -0.261717030
#> 11 15.242424 -0.329739301 -0.44963766 -0.209840943
#> 12 16.666667 -0.236456854 -0.33345593 -0.139457776
#> 13 18.090909 -0.187783201 -0.30292439 -0.072642015
#> 14 19.515152 -0.226748006 -0.33476704 -0.118728976
#> 15 20.939394 -0.229175092 -0.33080619 -0.127543993
#> 16 22.363636 -0.240824195 -0.36025925 -0.121389135
#> 17 23.787879 -0.396329451 -0.49288877 -0.299770132
#> 18 25.212121 -0.472299770 -0.58853860 -0.356060937
#> 19 26.636364 -0.346859356 -0.45344989 -0.240268819
#> 20 28.060606 -0.352051750 -0.45504127 -0.249062233
#> 21 29.484848 -0.509917727 -0.62863393 -0.391201528
#> 22 30.909091 -0.553447782 -0.64976105 -0.457134516
#> 23 32.333333 -0.458669461 -0.57595336 -0.341385561
#> 24 33.757576 -0.319163400 -0.42430868 -0.214018119
#> 25 35.181818 -0.274962044 -0.37937956 -0.170544531
#> 26 36.606061 -0.343014587 -0.46081001 -0.225219168
#> 27 38.030303 -0.415401411 -0.51166773 -0.319135092
#> 28 39.454545 -0.397422379 -0.51569997 -0.279144793
#> 29 40.878788 -0.285716394 -0.38941287 -0.182019919
#> 30 42.303030 -0.281039407 -0.38691070 -0.175168109
#> 31 43.727273 -0.364210514 -0.48097224 -0.247448788
#> 32 45.151515 -0.295850803 -0.39226072 -0.199440886
#> 33 46.575758 -0.202393200 -0.32149651 -0.083289890
#> 34 48.000000 -0.258064413 -0.36036400 -0.155764823
#> 35 49.424242 -0.294762948 -0.40206745 -0.187458442
#> 36 50.848485 -0.243743271 -0.35944671 -0.128039829
#> 37 52.272727 -0.212056003 -0.30881412 -0.115297886
#> 38 53.696970 -0.223624912 -0.34332922 -0.103920599
#> 39 55.121212 -0.245821845 -0.34682625 -0.144817439
#> 40 56.545455 -0.216226869 -0.32497022 -0.107483516
#> 41 57.969697 -0.122563126 -0.23710176 -0.008024492
#> 42 59.393939 -0.022064966 -0.11936732 0.075237389
#> 43 60.818182 0.043246936 -0.07680916 0.163303035
#> 44 62.242424 0.058536017 -0.04133763 0.158409663
#> 45 63.666667 0.064177908 -0.04607577 0.174431583
#> 46 65.090909 0.064875182 -0.04834389 0.178094259
#> 47 66.515152 0.005296521 -0.09272732 0.103320363
#> 48 67.939394 -0.105963819 -0.22614647 0.014218833
#> 49 69.363636 -0.206727445 -0.30561597 -0.107838917
#> 50 70.787879 -0.189575482 -0.30134938 -0.077801580
#> 51 72.212121 -0.076046167 -0.18782007 0.035727734
#> 52 73.636364 -0.079103697 -0.17799222 0.019784831
#> 53 75.060606 -0.162040947 -0.28222360 -0.041858296
#> 54 76.484848 -0.159214768 -0.25723861 -0.061190926
#> 55 77.909091 -0.096978598 -0.21019767 0.016240479
#> 56 79.333333 -0.034892264 -0.14514594 0.075361411
#> 57 80.757576 -0.001654978 -0.10152862 0.098218668
#> 58 82.181818 -0.016382605 -0.13643870 0.103673494
#> 59 83.606061 -0.082110591 -0.17941295 0.015191763
#> 60 85.030303 -0.149307173 -0.26384581 -0.034768540
#> 61 86.454545 -0.164889347 -0.27363270 -0.056145994
#> 62 87.878788 -0.103394785 -0.20439919 -0.002390379
#> 63 89.303030 -0.035126676 -0.15483099 0.084577636
#> 64 90.727273 -0.062581265 -0.15933938 0.034176852
#> 65 92.151515 -0.079086931 -0.19479037 0.036616512
#> 66 93.575758 -0.021958536 -0.12926304 0.085345970
#> 67 95.000000 -0.042156899 -0.14445649 0.060142691
#> 68 96.424242 -0.106105596 -0.22520891 0.012997714
#> 69 97.848485 -0.041483460 -0.13789338 0.054926457
#> 70 99.272727 0.086897127 -0.02986460 0.203658853
#> 71 100.696970 0.174316765 0.06844547 0.280188063
#> 72 102.121212 0.217565731 0.11386926 0.321262206
#> 73 103.545455 0.202671643 0.08439406 0.320949227
#> 74 104.969697 0.115659933 0.01939361 0.211926256
#> 75 106.393939 0.087320078 -0.03047534 0.205115498
#> 76 107.818182 0.208210642 0.10379313 0.312628152
#> 77 109.242424 0.335094675 0.22994939 0.440239956
#> 78 110.666667 0.360344269 0.24306036 0.477628175
#> 79 112.090909 0.268794480 0.17248123 0.365107728
#> 80 113.515152 0.189627094 0.07091091 0.308343273
#> 81 114.939394 0.240639673 0.13765014 0.343629210
#> 82 116.363636 0.346623508 0.24003297 0.453214045
#> 83 117.787879 0.413779514 0.29754069 0.530018341
#> 84 119.212121 0.384577682 0.28801824 0.481137123
#> 85 120.636364 0.369334466 0.24989923 0.488769704
#> 86 122.060606 0.473909701 0.37227863 0.575540771
#> 87 123.484848 0.560667717 0.45264868 0.668686751
#> 88 124.909091 0.568661309 0.45351981 0.683802810
#> 89 126.333333 0.576904017 0.47990550 0.673902532
#> 90 127.757576 0.592069051 0.47217142 0.711966684
#> 91 129.181818 0.589034350 0.48862971 0.689438988
#> 92 130.606061 0.577041729 0.46755078 0.686532679
#> 93 132.030303 0.575296888 0.46156731 0.689026471
#> 94 133.454545 0.606978315 0.50938891 0.704567721
#> 95 134.878788 0.696587825 0.57665900 0.816516648
#> 96 136.303030 0.838493153 0.74033856 0.936647746
#> 97 137.727273 0.917933747 0.80690816 1.028959338
#> 98 139.151515 0.894514412 0.78638554 1.002643279
#> 99 140.575758 0.865726591 0.76474733 0.966705850
#> 100 142.000000 0.859592517 0.68831338 1.030871657
#>
summary(model)
#> ---------------------------------------------------------------
#> Additive partial linear models with symmetric errors
#> ---------------------------------------------------------------
#> Sample size: 142
#> -------------------------- Model ---------------------------
#>
#> aplms::aplms(formula = temperature ~ 1, npc = c("time"), basis = c("cr"),
#> Knot = c(60), data = temperature.df, family = Powerexp(k = 0.3),
#> p = 1, control = list(tol = 0.001, algorithm1 = c("P-GAM"),
#> algorithm2 = c("BFGS"), Maxiter1 = 20, Maxiter2 = 25),
#> lam = c(10))
#>
#> ------------------- Parametric component -------------------
#>
#> Estimate Std. Error t value Pr(>|t|)
#> intercept 0.056619 0.0041 13.8905 < 2.2e-16 ***
#>
#> ----------------- Non-parametric component ------------------
#>
#> Wald df Pr(>.)
#> time 7589.838 58.583 < 2.2e-16 ***
#>
#> --------------- Autoregressive and Scale parameter ----------------
#>
#> Estimate Std. Error Wald Pr(>|t|)
#> phi 0.0022992 0.0003 7.3902 1.242e-11 ***
#> rho1 -0.2571215 0.0662 -3.8866 0.0001568 ***
#>
#>
#> ------ Penalized Log-likelihood and Information criterion------
#>
#> Log-lik: 200.07
#> AIC : -276.97
#> AICc : -274.46
#> BIC : -94.94
#> GCV : 0.01
#>
#> --------------------------------------------------------------------
print(model)
#> Call:
#> aplms::aplms(formula = temperature ~ 1, npc = c("time"), basis = c("cr"),
#> Knot = c(60), data = temperature.df, family = Powerexp(k = 0.3),
#> p = 1, control = list(tol = 0.001, algorithm1 = c("P-GAM"),
#> algorithm2 = c("BFGS"), Maxiter1 = 20, Maxiter2 = 25),
#> lam = c(10))